Originally Posted by apredeus PS sorry - you already got the question answered, I'm still asleep apparently. Starting over is never fun. Powered by Biostar version Hi, I have the following bp reads. Is there a way to extra 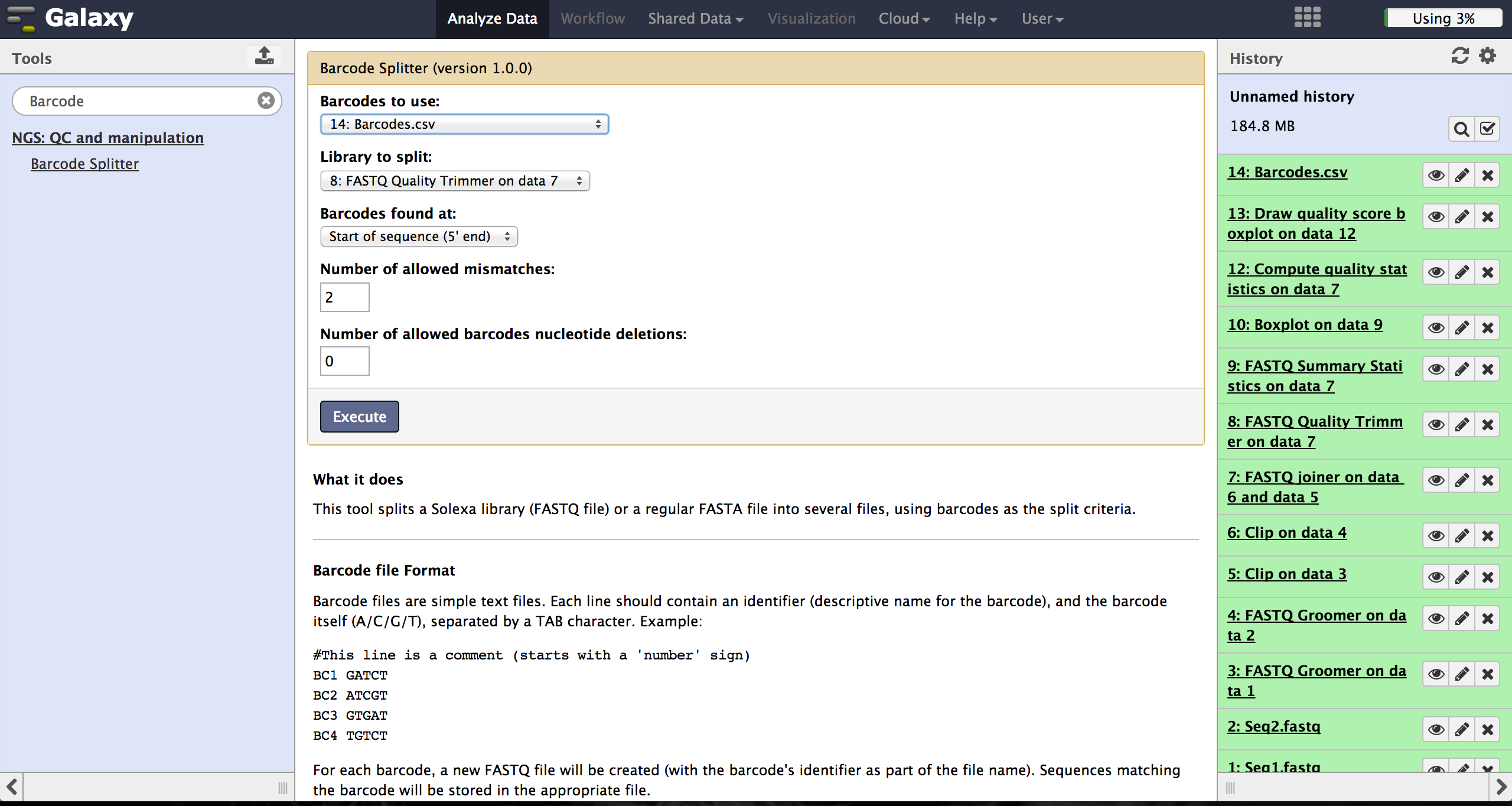
| Uploader: | Vudobei |
| Date Added: | 24 September 2009 |
| File Size: | 61.66 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 29027 |
| Price: | Free* [*Free Regsitration Required] |
Any recommendations for faster splitting? Originally Posted by ehlin I haven't tried this, but would it be faster if you specified a file size rather than a line number? When using this tool it fails to recognize my data files for input.
FASTQ Splitter
The script won't cut any sequence in the middle, or change the order of sequences. I have to analyze paired-end RNA-seq read that are in an unusual format: I'm new to bioinformatics but based on the encoding values I still have no idea what I should be using. Split fastq file with renaming reads. So, dataset format to specification and consistent content are the primary components you want to focus on at first, to meet tool input requirements.
Find More Posts by gsgs.
Fastq Splitter For Paired End Reads
Filter SAM Tool version: Apologies if this has been asked before. Limitations Wrapped multi-line sequence and quality is currently not supported. There's a blog splitteg on how you can use this utility to create a.
I believe Splittee have an illumina 1. I haven't tried this, but would it be faster if you specified a file size rather than a line number?
I want to do the mapping about AB S I want to sort my fastq Last edited by pallevillesen; at I don't know fasstq the mapping procedure doesn't work. It allows processing the dataset in parallel, for instance on a cluster computer. Now I'm trying to do the ex I had to convert my fastq into wplitter in order to t It is shared with the hope that it can be useful, but without any warranties.
Now need to use Pindel split-read approach to identify CNV. User support for Galaxy!
Use of this site constitutes acceptance of our User Agreement and Privacy Policy. Hi, I am very happy to try the software on http: Splits a single fastq dataset representing paired-end run into two datasets one for spltiter end. Berlin, DE Join Date: Send a private message to pallevillesen. In that case, there is a nice option: Find More Posts by apredeus. Originally Posted by pallevillesen.
Very elegant, but string indices in awk are splitfer so that as is, above code duplicates the last base of the first read as the first base of the second read. Hi, How can I merge two paired end fastq R and L to give a single fastq file? It instead downloads files from SRA and converts them into fastq files for you. Send a private message to apredeus.

Comments
Post a Comment